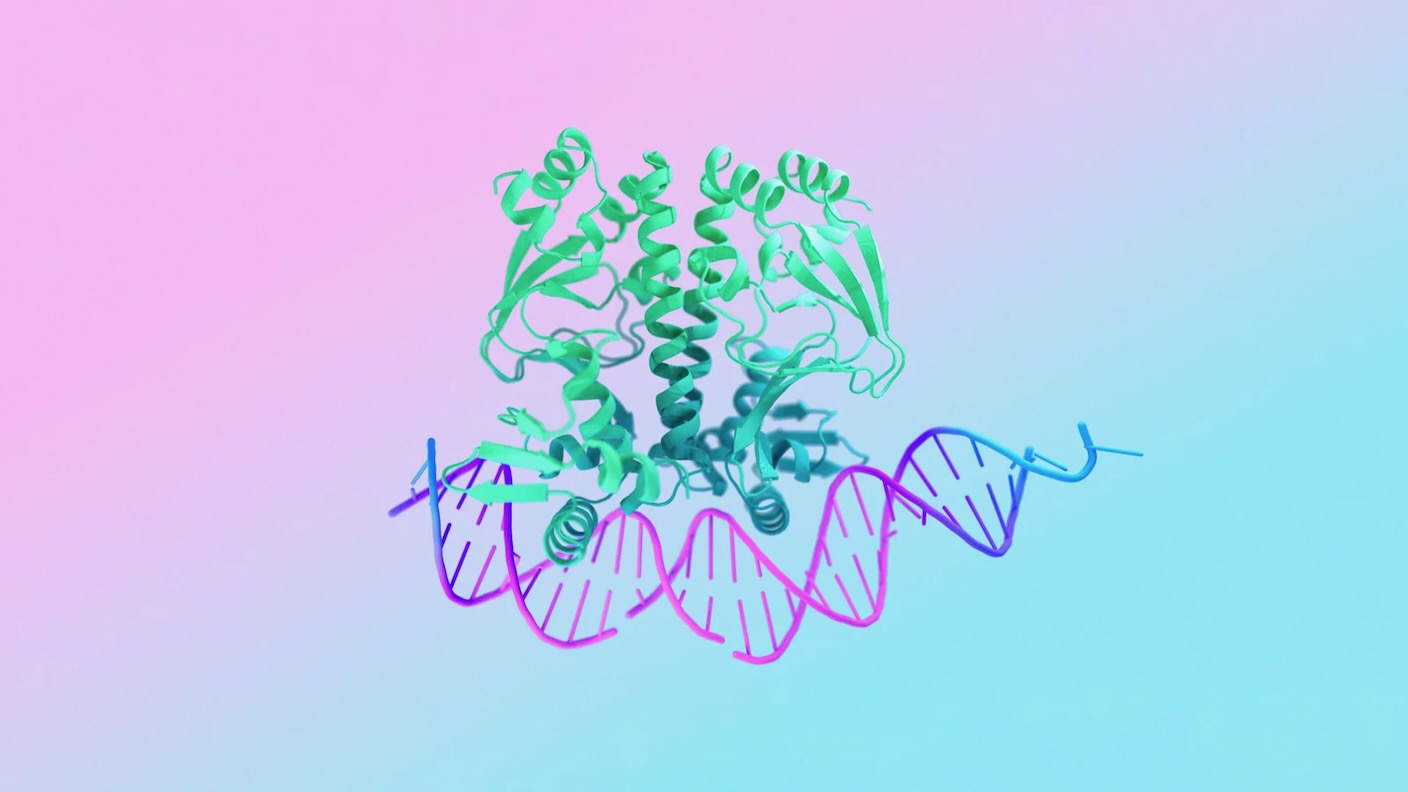
Proteins are organic workhorses.
They construct our our bodies and orchestrate the molecular processes in cells that hold them wholesome. In addition they current a wealth of targets for brand new medicines. From on a regular basis ache relievers to stylish most cancers immunotherapies, most present medication work together with a protein. Deciphering protein architectures might result in new remedies.
That was the promise of AlphaFold 2, an AI mannequin from Google DeepMind that predicted how proteins acquire their distinctive shapes based mostly on the sequences of their constituent molecules alone. Launched in 2020, the instrument was a breakthrough half a decade within the making.
However proteins don’t work alone. They inhabit a complete mobile universe and sometimes collaborate with different molecular inhabitants like, for instance, DNA, the physique’s genetic blueprint.
This week, DeepMind and Isomorphic Labs launched a giant new replace that enables the algorithm to foretell how proteins work inside cells. As an alternative of solely modeling their constructions, the brand new model—dubbed AlphaFold 3—may also map a protein’s interactions with different molecules.
For instance, might a protein bind to a disease-causing gene and shut it down? Can including new genes to crops make them resilient to viruses? Can the algorithm assist us quickly engineer new vaccines to deal with present ailments—or no matter new ones nature throws at us?
“Biology is a dynamic system…it’s important to perceive how properties of biology emerge as a result of interactions between completely different molecules within the cell,” stated Demis Hassabis, the CEO of DeepMind, in a press convention.
AlphaFold 3 helps clarify “not solely how proteins speak to themselves, but in addition how they speak to different elements of the physique,” stated lead writer Dr. John Jumper.
The group is releasing the brand new AI on-line for tutorial researchers by means of an interface known as the AlphaFold Server. With a number of clicks, a biologist can run a simulation of an thought in minutes, in comparison with the weeks or months normally wanted for experiments in a lab.
Dr. Julien Bergeron at King’s School London, who builds nano-protein machines however was not concerned within the work, stated the AI is “transformative science” for rushing up analysis, which might finally result in nanotech units powered by the physique’s mechanisms alone.
For Dr. Frank Uhlmann on the Francis Crick Laboratory, who gained early entry to AlphaFold 3 and used it to check how DNA divides when cells divide, the AI is “democratizing discovery analysis.”
Molecular Universe
Proteins are finicky creatures. They’re manufactured from strings of molecules known as amino acids that fold into intricate three-dimensional shapes that decide what the protein can do.
Typically the folding processes goes unsuitable. In Alzheimer’s illness, misfolded proteins clump into dysfunctional blobs that clog up round and inside mind cells.
Scientists have lengthy tried to engineer medication to interrupt up disease-causing proteins. One technique is to map protein construction—know thy enemy (and associates). Earlier than AlphaFold, this was accomplished with electron microscopy, which captures a protein’s construction on the atomic stage. However it’s costly, labor intensive, and never all proteins can tolerate the scan.
Which is why AlphaFold 2 was revolutionary. Utilizing amino acid sequences alone—the constituent molecules that make up proteins—the algorithm might predict a protein’s remaining construction with startling accuracy. DeepMind used AlphaFold to map the construction of practically all proteins identified to science and the way they work together. In accordance with the AI lab, in simply three years, researchers have mapped roughly six million protein constructions utilizing AlphaFold 2.
However to Jumper, modeling proteins isn’t sufficient. To design new medication, it’s important to assume holistically in regards to the cell’s complete ecosystem.
It’s an thought championed by Dr. David Baker on the College of Washington, one other pioneer within the protein-prediction area. In 2021, Baker’s group launched AI-based software program known as RoseTTAFold All-Atom to deal with interactions between proteins and different biomolecules.
Picturing these interactions may help clear up powerful medical challenges, permitting scientists to design higher most cancers remedies or extra exact gene therapies, for instance.
“Properties of biology emerge by way of the interactions between completely different molecules within the cell,” stated Hassabis within the press convention. “You may take into consideration AlphaFold 3 as our first massive kind of step in direction of that.”
A Revamp
AlphaFold 3 builds on its predecessor, however with important renovations.
One option to gauge how a protein interacts with different molecules is to look at evolution. One other is to map a protein’s 3D construction and—with a dose of physics—predict the way it can seize onto different molecules. Whereas AlphaFold 2 principally used an evolutionary method—coaching the AI on what we already learn about protein evolution in nature—the brand new model closely embraces bodily and chemical modeling.
A few of this contains chemical adjustments. Proteins are sometimes tagged with completely different chemical substances. These tags generally change protein construction however are important to their habits—they’ll actually decide a cell’s destiny, for instance, life, senescence, or loss of life.
The algorithm’s total setup makes some use of its predecessor’s equipment to map proteins, DNA, and different molecules and their interactions. However the group additionally regarded to diffusion fashions—the algorithms behind OpenAI’s DALL-E 2 picture generator—to seize constructions on the atomic stage. Diffusion fashions are educated to reverse noisy pictures in steps till they arrive at a prediction for what the picture (or on this case a 3D mannequin of a biomolecule) ought to appear to be with out the noise. This addition made a “substantial change” to efficiency, stated Jumper.
Like AlphaFold 2, the brand new model has a built-in “sanity examine” that signifies how assured it’s in a generated mannequin so scientists can proofread its outputs. This has been a core part of all their work, stated the DeepMind group. They educated the AI utilizing the Protein Knowledge Financial institution, an open-source compilation of 3D protein constructions that’s always up to date, together with new experimentally validated constructions of proteins binding to DNA and different biomolecules
Pitted towards present software program, AlphaFold 3 broke information. One take a look at for molecular interactions between proteins and small molecules—ones that might change into medicines—succeeded 76 p.c of the time. Earlier makes an attempt have been profitable in roughly 42 p.c of instances.
In terms of deciphering protein features, AlphaFold 3 “seeks to resolve the very same drawback [as RoseTTAFold All-Atom]…however is clearly extra correct,” Baker instructed Singularity Hub.
However the instrument’s accuracy relies on which interplay is being modeled. The algorithm isn’t but nice at protein-RNA interactions, for instance, Columbia College’s Mohammed AlQuraishi instructed MIT Know-how Evaluate. Total, accuracy ranged from 40 to greater than 80 p.c.
AI to Actual Life
Not like earlier iterations, DeepMind isn’t open-sourcing AlphaFold 3’s code. As an alternative, they’re releasing the instrument as a free on-line platform, known as AlphaFold Server, that enables scientists to check their concepts for protein interactions with only a few clicks.
AlphaFold 2 required technical experience to put in and run the software program. The server, in distinction, may help individuals unfamiliar with code to make use of the instrument. It’s for non-commercial use solely and might’t be reused to coach different machine studying fashions for protein prediction. However it’s freely accessible for scientists to strive. The group envisions the software program serving to develop new antibodies and different remedies at a sooner fee. Isomorphic Labs, a spin-off of DeepMind, is already utilizing AlphaFold 3 to develop medicines for quite a lot of ailments.
For Bergeron, the improve is “transformative.” As an alternative of spending years within the lab, it’s now attainable to imitate protein interactions in silico—a pc simulation—earlier than starting the labor- and time-intensive work of investigating promising options utilizing cells.
“I’m fairly sure that each structural biology and protein biochemistry analysis group on the earth will instantly undertake this technique,” he stated.
Picture Credit score: Google DeepMind