Cells talk with their environments through the plasma membrane and varied membrane proteins. Clathrin-mediated endocytosis (CME) performs a central function in such communication and proceeds with a sequence of multiprotein meeting, deformation of the plasma membrane, and manufacturing of a membrane vesicle that delivers extracellular signaling molecules into the cytoplasm.*
Within the article “Morphological adjustments of plasma membrane and protein meeting throughout clathrin-mediated endocytosis”, Aiko Yoshida, Nobuaki Sakai, Yoshitsugu Uekusa, Yuka Imaoka, Yoshitsuna Itagaki, Yuki Suzuki and Shige H. Yoshimura describe how they utilized their home-built correlative imaging system comprising high-speed atomic drive microscopy (HS-AFM) and confocal fluorescence microscopy to concurrently picture morphological adjustments of the plasma membrane and protein localization throughout CME in a residing cell.*
Overlaying AFM and fluorescence photographs revealed the dynamics of protein meeting and concomitant morphological adjustments of the plasma membrane with excessive spatial decision. Specifically, the authors elucidate the function of actin within the closing step of CME.*
The outcomes revealed a decent correlation between the dimensions of the pit and the quantity of clathrin assembled. Actin dynamics play a number of roles within the meeting, maturation, and shutting phases of the method, and impacts membrane morphology, suggesting an in depth relationship between endocytosis and dynamic occasions on the cell cortex. Knock down of dynamin additionally affected the closing movement of the pit and confirmed practical correlation with actin.*
An AFM tip-scan–kind HS-AFM unit mixed with an inverted fluorescence/optical microscope geared up with a section distinction system and a confocal unit was used for this examine.*
The modulation technique was set to section modulation mode to detect AFM tip–pattern interactions. A custom-made NanoWorld Extremely-Brief AFM cantilever with an electron beam–deposited sharp AFM tip with a spring fixed of 0.1 N m−1 (USC-F0.8-k0.1-T12) was used. *
All observations have been carried out at 28 °C. The AFM tip was aligned with confocal views as described within the Outcomes part of the article. The pictures from the confocal microscope and AFM have been concurrently acquired at a scanning fee of 10 s/body. The captured sequential photographs have been overlaid by utilizing AviUTL (http://spring-fragrance.mints.ne.jp/aviutl/) based mostly on the AFM tip place.
The fluorescence depth was quantified by Picture J software program (http://rsbweb.nih.gov/ij/). *
Aligning the confocal picture and the AFM picture.
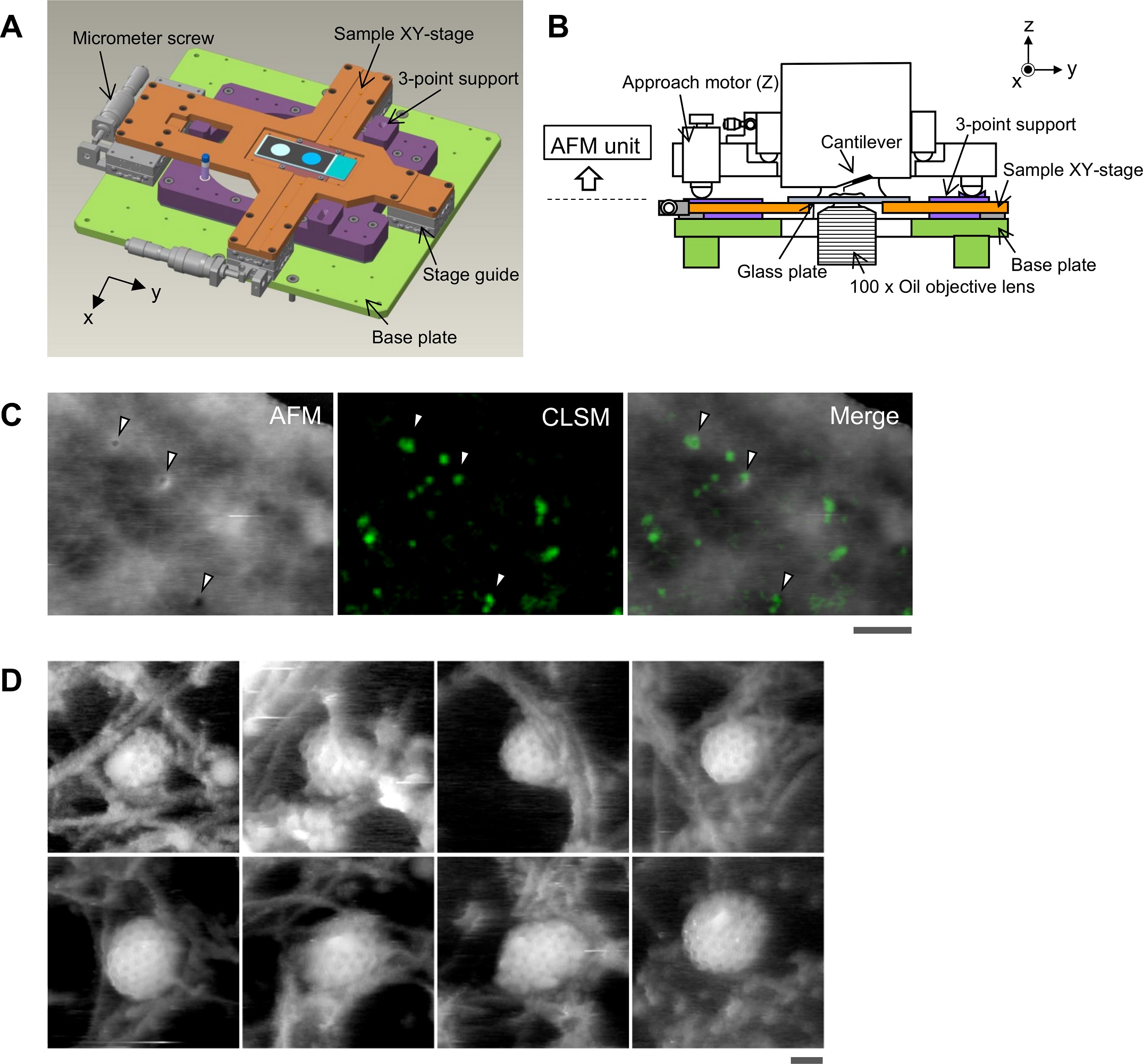
(A) Schematic illustration of the pattern stage. A cross-shaped movable XY-stage (orange) is mounted on the bottom plate (gentle inexperienced) of the inverted optical microscope (IX83) through a stage information (grey) geared up at every of the 4 ends of the cross. A 3-point assist plate (purple) for mounting the AFM scanner unit is mounted on the bottom plate with a configuration that doesn’t hinder the sliding of the XY-stage alongside the x-axis and y-axis. These setups enable the pattern stage to maneuver independently of the AFM unit and the optical axis. (B) Aspect view of the HS-AFM unit mounted on the stage illustrated in panel A. (C) Overlaying a confocal picture and an AFM picture. COS-7 cells expressing EGFP-CLCa have been mounted with 5% paraformaldehyde and subjected to AFM (left) and CLSM (center) imaging. The x-y place of the probe tip was decided as described in S1 Fig. Two photographs have been overlaid (proper) based mostly on the x-y middle place. Scale bar: 1 μm. Autofluorescence of the probe was a lot weaker than clathrin spot and couldn’t be detected in the course of the quick scanning. (D) AFM photographs of CCP on the cytoplasmic floor of the plasma membrane. COS-7 cells have been “unroofed” by gentle sonication as described in Supplies and strategies after which mounted with glutaraldehyde. Scale bar: 0.1 μm. AFM, atomic drive microscopy; CCP, clathrin-coated pit; CLSM, confocal laser scanning microscopy; COS-7, CV-1 in origin with SV40 gene line 7; EGFP, enhanced inexperienced fluorescent protein; EGFP-CLCa, EGFP-fused clathrin gentle chain a; HS-AFM, high-speed AFM.
https://doi.org/10.1371/journal.pbio.2004786.g001
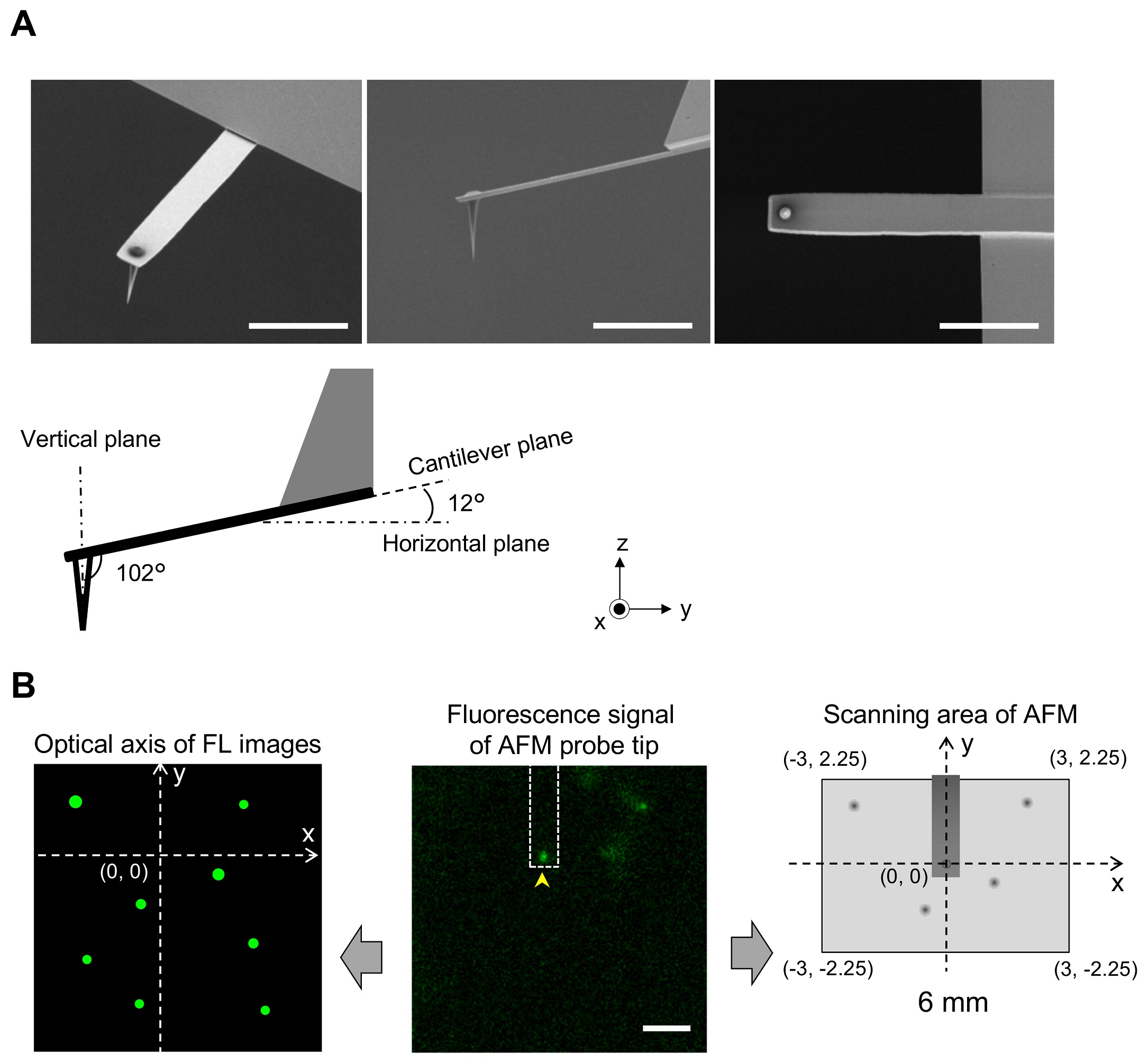
1 Fig. Aligning confocal and AFM photographs.
(A) Scanning electron microscopy (SEM) photographs of a cantilever geared up with an EBD tip with tilt angle of 12°. Scale bar, 5 μm. Be aware that the cantilever is held on the AFM head unit with a tilt angle of 102° (from the x-y airplane) in order that the relative tip–pattern angle (θ) is 90°. This setup makes it doable to exactly decide the place of the AFM tip. Scale bar, 2 μm. (B) Figuring out the place of the AFM probe in a fluorescence picture. Whereas the AFM probe was hooked up on the glass floor with out scanning, the autofluorescence sign of the probe was imaged by the confocal scanning unit. The noticed fluorescence spot (arrowhead within the center panel) is outlined as an origin of the fluorescence picture airplane (x = 0, y = 0) and used to outline the optical axis (left panel). The place of a fluorescence spot derived from EGFP-CLCa was decided on this axis. Alternatively, the scanning space of the AFM scanner covers the world of (−3, 2.25) (left prime), (3, 2.25) (proper prime), (3, −2.25) (proper backside), and (−3, −2.25) (left backside) (all proper panel). By aligning the axis from each photographs, the x, y place of the AFM picture and that of the confocal fluorescence picture may very well be merged. AFM, atomic drive microscopy; EBD, electron beam–deposited; EGFP, enhanced inexperienced fluorescent protein; EGFP-CLCa, EGFP-fused clathrin gentle chain a.
https://doi.org/10.1371/journal.pbio.2004786.s001
*Aiko Yoshida, Nobuaki Sakai, Yoshitsugu Uekusa, Yuka Imaoka, Yoshitsuna Itagaki, Yuki Suzuki and Shige H. Yoshimura
Morphological adjustments of plasma membrane and protein meeting throughout clathrin-mediated endocytosis
PLoS Biol 16(5) (2018): e2004786
DOI: https://doi.org/10.1371/journal.pbio.2004786
The article “Morphological adjustments of plasma membrane and protein meeting throughout clathrin-mediated endocytosis” by Aiko Yoshida, Nobuaki Sakai, Yoshitsugu Uekusa, Yuka Imaoka, Yoshitsuna Itagaki, Yuki Suzuki and Shige H. Yoshimura is licensed below a Inventive Commons Attribution 4.0 Worldwide License, which allows use, sharing, adaptation, distribution and replica in any medium or format, so long as you give acceptable credit score to the unique writer(s) and the supply, present a hyperlink to the Inventive Commons license, and point out if adjustments have been made. The pictures or different third-party materials on this article are included within the article’s Inventive Commons license, until indicated in any other case in a credit score line to the fabric. If materials shouldn’t be included within the article’s Inventive Commons license and your supposed use shouldn’t be permitted by statutory regulation or exceeds the permitted use, you have to to acquire permission straight from the copyright holder. To view a duplicate of this license, go to https://creativecommons.org/licenses/by/4.0/.

